Google’s DeepMind division used AlphaFold, an AI system it built internally, to make predictions about the structures of nearly all proteins currently known to science. Today, DeepMind revealed that it had reached a significant milestone.
Scientists can access the protein structures predicted by Alphabet’s artificial intelligence system through UniProt, a freely available data repository for proteins. DeepMind expects the data to speed up new drug discovery and enhance research in a variety of other fields as well. DeepMind.
Read More-
- Debris From A Chinese Rocket Is Crashing Toward Earth Could Fall In Next Few Days!
- Meta Quest 2 VR Headset Price Increase Announced Is Getting A $100 On August 1?
Amino acids are the building blocks of proteins, which are the building blocks of life. As many as twenty different amino acids interact in a variety of ways. When amino acids interact, proteins take on a three-dimensional structure that has long been the interest of scientists.
A protein’s behavior is directly influenced by its structure. This means that determining the structure of a protein is a critical first step in many studies. With AlphaFold, DeepMind has created an AI system that will aid scientists in their work.
Amino acid organization in a protein can be determined using a variety of techniques. The downside is that these approaches take a long time to complete: According to DeepMind, it can take years to fully understand the structure of a single protein.
DeepMind’s AlphaFold tool has determined the structures of around 200 million proteins, from almost every known organism on Earth. https://t.co/Zrhj5Io2Uv
— AnnaEsse (@AnnaEsse) July 28, 2022
Furthermore, the task requires a large amount of expensive equipment. It’s taken decades of research for scientists to build algorithms that can predict the shape of a protein based on the amino acids it contains. It has been difficult to design software for this activity because of its intricacy.
There are so many possible configurations in a typical protein, DeepMind estimates, that calculating them one by one would take longer than the cosmos has been around. That’s not a problem for DeepMind’s AlphaFold. AlphaFold was first described by the Alphabet unit in 2018 and a fresh version of the AI system with enhanced capabilities was revealed two years later.
Protein structure predictions were made with an average error rate of roughly 1.6 angstroms or the width of one atom. AlphaGo has helped DeepMind anticipate more than 200 million protein structures. Most proteins known to science have been predicted by the Alphabet unit.
“This update includes predicted structures for plants, bacteria, animals, and other organisms, opening up many new opportunities for researchers to use AlphaFold to advance their work on important issues, including sustainability, food insecurity, and neglected diseases,” DeepMind Chief Executive Officer Demis Hassabis wrote in a blog post.
Using the UniProt protein database, DeepMind is making its predicted protein structures dataset available to the general public. In addition, the datasets will be made available as open-source datasets on Google Cloud. One million predicted protein structures disclosed previously by DeepMind have already been used by more than 500,000 researchers, according to the company.
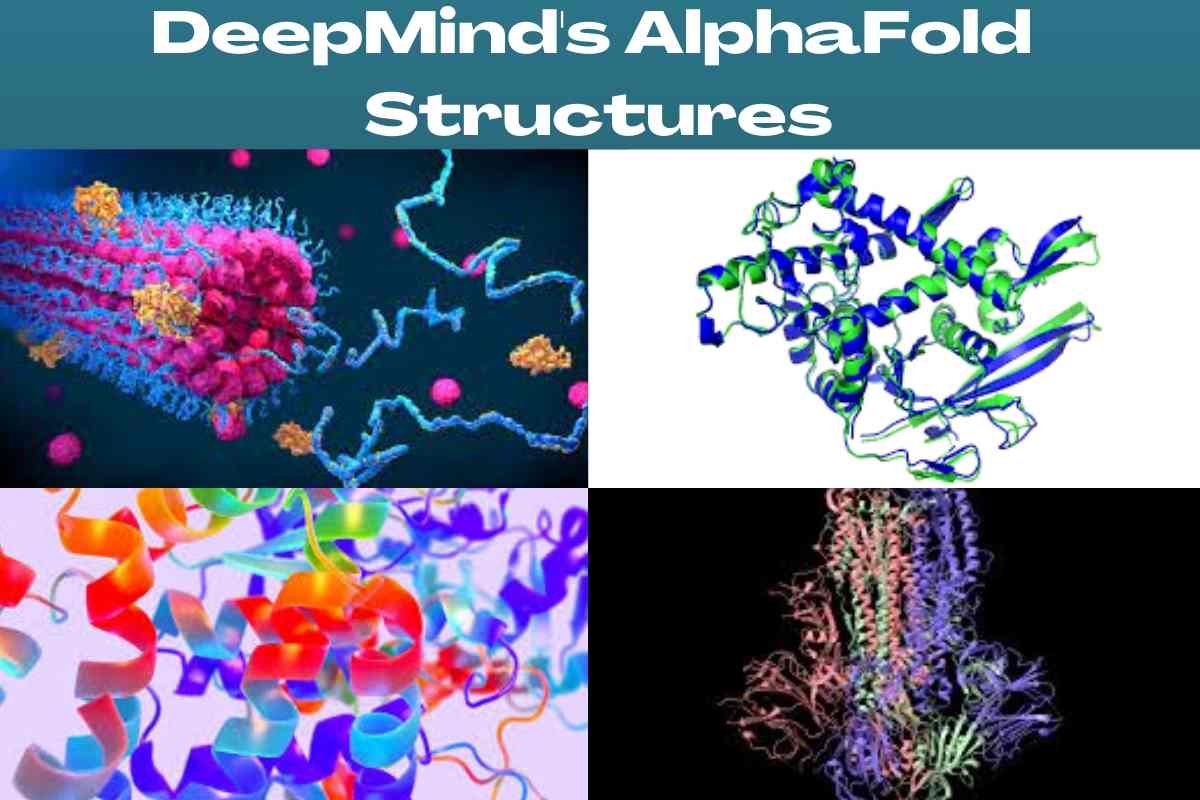
It is only the beginning of the impact that we will start to witness over the coming few years, Hassabis stated, “from combating disease to producing vaccines, AlphaFold has already enabled amazing advancements on some of our biggest global concerns”. When applied to biology, “DeepMind’s AlphaFold represents a peek of the future and what can be achieved with computational and AI technologies.”
DeepMind is also looking into new uses for AI in other industries. Machine learning systems that can play Go and predict the weather have been built by the research team. Since then, DeepMind has released a system for repairing ancient Greek writings, as well as determining when they were written.




